Specific steps of Edman degradation
Edman degradation is a very important reaction step in protein sequencing, because the ordered amino acid composition of protein can be analyzed through this reaction. Automated Edman sequencing machines are now widely used, capable of sequencing peptides that are approximately 50 amino acids long. The steps of protein sequencing using Edman degradation are summarized below, and some of the important steps will be described in detail later.
1. Use a reducing agent such as 2-mercaptoethanol to destroy any disulfide bond in the protein, and a protective group (such as iodoacetic acid) may be required in the process to prevent the chemical bond from re forming;
2. If there is more than one protein, it is necessary to separate and purify each peptide chain in the protein mixture;
3. Determine the amino acid composition in each chain;
4. Determine the amino acids at the end of each chain;
5. Break each chain into fragments of less than 50 amino acids;
6. Isolation and purification of peptide fragments;
7. Determine the sequence of each peptide fragment;
8. Repeat the above steps using other cracking methods;
9. Construct the entire protein sequence
The process of protein cleavage into peptide fragments
Peptides longer than 50-70 amino acids cannot be sequenced by Edman degradation. Therefore, it is necessary to break down long protein chains into small fragments and then perform separate sequencing. The lysis process can be carried out through endopeptidases (such as trypsin or pepsin) or chemical reagents (such as cyanide bromide). Different enzymes have different cleavage modes, and the overlap between fragments can be used to construct overall protein sequences.
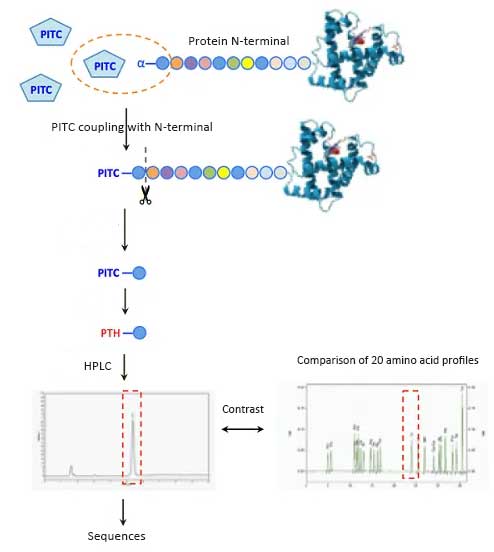
Edman reaction process
Adsorb the protein to be sequenced onto a solid surface. A common substrate is glass fiber coated with polybutadiene, a cationic polymer. Edman reagent phenyl isothiocyanate (PITC) and 12% trimethylamine weak base buffer solution were added to the adsorbed protein, and the product was reacted with the amino group at the N-terminal of amino acid.
Then, the terminal amino acids are selectively separated by adding anhydrous acid. Next, the derivative undergoes isomerization to obtain the substitute phenyl thiohydantoin. Wash off the substitute and identify it by chromatography, repeating the entire cycle. The efficiency of each step is about 98%, which can effectively identify about 50 amino acids.
The Use of Protein Sequencers
The protein sequencer is a machine that performs Edman degradation in an automated manner. Fix the sample of protein or peptide in the reaction container of protein sequencer for Edman degradation. Each cycle releases and derives an amino acid from the N-terminal of a protein or peptide, and then identifies the resulting amino acid derivative through HPLC. Repeat the sequencing process for the entire peptide until the complete measurable sequence or predetermined number of cycles have been determined.
>> Custom peptide synthesis company China - Omizzur
Copyright © 2020 Omizzur Inc | Terms & Conditions | Privacy Notice | Sitemap