Peptide Characterization APIs(1): Identification
The molecular mass of peptides is between typical organic small molecules and high molecular compounds. In general, a peptide is a compound composed of ≤ 45 amino acids linked by peptide bonds. Peptides widely exist in the human body and have important physiological effects, and peptide drugs have long been used in the clinical treatment of diabetes and certain cancers.
With the continuous emergence of peptide drugs, the importance of peptide-related analysis and detection research has become increasingly prominent. In the production of peptide raw materials in pharmaceutical companies, if there is no strict analysis, characterization and impurity detection, the product will have problems and may cause very serious consequences.
The drug quality control department is an important part of drug quality , and the product can only be released if the test results meet the pre-established indicators. With the increasing number of peptide drugs on the market, drug regulatory agencies must establish quality control parameters for various peptide APIs.
In order to ensure the high quality and consistency of the manufactured drug product, the analysis and characterization of the peptide drug substance must consider several quality control parameters to ensure that the pharmaceutical quality assurance department can fully evaluate the quality of the product before releasing it
The quality of peptide raw materials is closely related to its production process, and even if the products originate from the same production process, the quality of products from different production batches may be different. Therefore, the structural integrity characterization and quality testing of peptide APIs are crucial. The following describes the analysis and characterization of peptide APIs from the aspects of identification, inspection (purity), content determination and other general attribute testing.
Peptide characterization 1: Identification of Peptide
Identification is mainly to confirm the structure of the peptide, that is, to confirm the identity of the product. The identification of peptides is not simple and can be completed by a single detection. The confirmation of its primary structure generally uses mass spectrometry to determine the relative molecular mass, chromatography to determine the amino acid composition, and mass spectrometry combined with enzyme digestion or Edman degradation to determine the peptide sequence.
Edman degradation is a traditional and effective peptide sequence determination method, but it is generally not suitable for peptides with closed or cyclic amino terminals; the number of amino acids detected by mass spectrometry should not exceed 25, otherwise it will cause data loss and difficult to parse.
The secondary structure of a peptide is actually a local structure formed by the peptide chain due to hydrogen bonds. Its confirmation generally includes detection methods such as X-ray single crystal diffraction, nuclear magnetic resonance, infrared, ultraviolet, and circular dichroism. The identification of the peptide structure is commonly used and not limited to the above-mentioned detection methods. Since each detection method can only obtain some specific information of the peptide structure, the use of orthogonal peptide characterization methods is the key to ensure accurate identification of the peptide secondary structure.
Generally speaking, the structural characteristics of short peptides can be obtained by using magnetic resonance, infrared, ultraviolet detection and amino acid composition analysis, but for long peptides, amino acid sequence analysis combined with mass spectrometry detection is required to confirm the accuracy of the peptide sequence Sexuality, that is, more detection and analysis are required, and comprehensive analysis and attribution of the obtained information are required.
It must be pointed out that: if standard peptides can be obtained, the spectra of the peptides to be tested can be compared with the spectra of the standard, which greatly simplifies and facilitates the identification of peptides. Which detection indicators need to be set for the identification of peptides depends on the production process, chain length characteristics of the peptide and its matching degree with the detection method.
Omizzur ltd is custom peptide synthesis company in China ,for peptide impurity synthesis please mail us:
Remark:
Edman degradation:
Edman degradation is a very important reaction step in protein sequencing, because the primary structure of peptides can be identified through this reaction, and the amino acid composition of proteins can be analyzed. Edman degradation is the process of determining the sequence of amino acid residues from the free N-terminus of a peptide chain.
The N-terminal amino acid residues of the polypeptide are modified by phenyl isothiocyanate (PITC), and the N-terminal of the peptide chain is selectively cut off to obtain thiazolinone aniline derivatives of the N-terminal amino acid residues. The derivative is then extracted with an organic solvent to form a stable phenylthiohydantoin (PTH) derivative. Which amino acid it is can be identified by analyzing the produced phenylhydantoin (PTH-amino acid) by HPLC or electrophoresis. Each reaction results in a peptide with the N-terminal amino acid residue removed, and the remaining peptide chain can enter the next cycle and continue to be degraded.
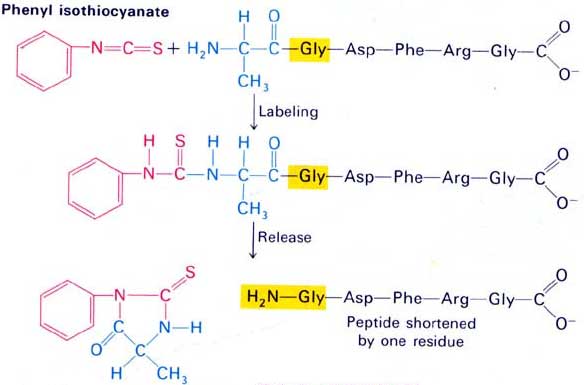
The above methods can already be automated. When the automatic analyzer designed according to this principle is used for analysis, the sequence of about 30 amino acid residues of the peptide chain can be reliably determined, and a maximum of 50-60 amino acid residues can be analyzed. Under the best conditions, the efficiency can be maintained above 99% for each formation of PTH-amino acid. Moreover, the amount of this method is small, and generally only 10-100 picomoles of peptide can be used to determine the amino acid sequence. For peptides with longer peptide chains, the peptide chain can be cut into multiple small peptides first, the amino acid analysis of these small peptides is carried out, and then the information is spliced together to obtain the amino acid sequence in the starting peptide chain.
Limitations of Edman's degradation method
Since Edman degradation is based on the modification of the N-terminal amino group by chemical reagents, when the N-terminal is blocked by other chemical groups, these groups need to be removed before sequencing. In addition, when a protein contains cysteine residues, two cysteine residues may be disulfide-crosslinked. In this case, the disulfide bond needs to be oxidized to –SO3H by peracid before sequence can be determined by Edman degradation.
Copyright © 2020 Omizzur Inc | Terms & Conditions | Privacy Notice | Sitemap