DNA-peptide complex molecule 2: self-assembly and regulation
Principle of self-assembly of DNA-peptide
Self-assembly is a process in which basic structural units (atoms, molecules, nanoparticles, etc.) are arranged in an orderly manner to form higher-scale structures through non-covalent interactions (weak interactions). The synergy of weak interaction forces (hydrogen bonds, van der Waals forces, electrostatic forces, hydrophobic forces, π-π stacking, cation π adsorption, etc.) between structural units is the driving force for self-assembly.
Peptides are chain compounds formed by connecting amino acids through peptide bonds. The hydrogen bonds between peptide bonds and various weak interactions between amino acids make them have the function of self-assembly. A linear sequence of amino acids connected to each other by peptide bonds is the primary structure of a peptide. Peptide self-assembly is generally based on its secondary structure, that is, the regular folding of polypeptide chain segments, and the common types are α-helix and β-sheet.
In a rod-shaped α-helix, the amino acids arrange themselves into a regular helical conformation (as shown in Figure 2a). The carbonyl oxygen of each peptide bond forms a hydrogen bond with a hydrogen far away from the amino group of the fourth amino acid, and the direction of the hydrogen bond is almost parallel to the helical axis. The positions of the amino acid side chains are located on the outside of the whole spirochete.
In the β-sheet, the planarity of the peptide bonds causes the polypeptide to fold into a sheet, and the amino acid side chains stretch on the upper and lower sides of the folded sheet (as shown in Figure 2b). In addition, the self-assembly of peptides also depends on their own amphiphilic structure. When amphiphilic short peptide molecules interact with water molecules, the hydrophilic segment will be adjacent to the water molecule, while the hydrophobic segment will gather inward (as shown in Figure 2c). Generally, amphiphilic peptides are constructed by combining hydrophilic peptide sequences with lipophilic groups, or by introducing hydrophobic and hydrophilic functional groups at the end of the peptide chain.
DNA consists of two intertwined strands forming a double helix structure with the base on the inside and the sugar-phosphate backbone on the outside. In a double helix, the two DNA strands are arranged antiparallel. The bases on the two strands pair complementary by hydrogen bonding. This highly predictable Watson-Crick base-pairing rule of DNA provides almost unlimited programmability for the synthesis of complex nanostructures.
In addition, through hybridization reactions and strand displacement reactions between DNAs, supramolecular assemblies with dynamically controllable structures and functions can also be constructed. Take the DNA strand displacement reaction as an example, which refers to the process in which two partially or fully complementary strands hybridize to each other, displacing one or more prehybridized strands (as shown in Figure 2d.
Strand displacement can initiate at complementary single-strand domains (called toe points) and proceed through a random walk-like branch migration process. This mechanism enables the kinetic control of the assembly path. Therefore, peptide-DNA composite molecules combine the biological functionality and assembly properties of peptides, the programmability and specificity of DNA, and can obtain multi-level ordered supramolecules that cannot be formed by simple peptides or DNA molecules through a variety of assembly regulation mechanisms and Functional Materials.
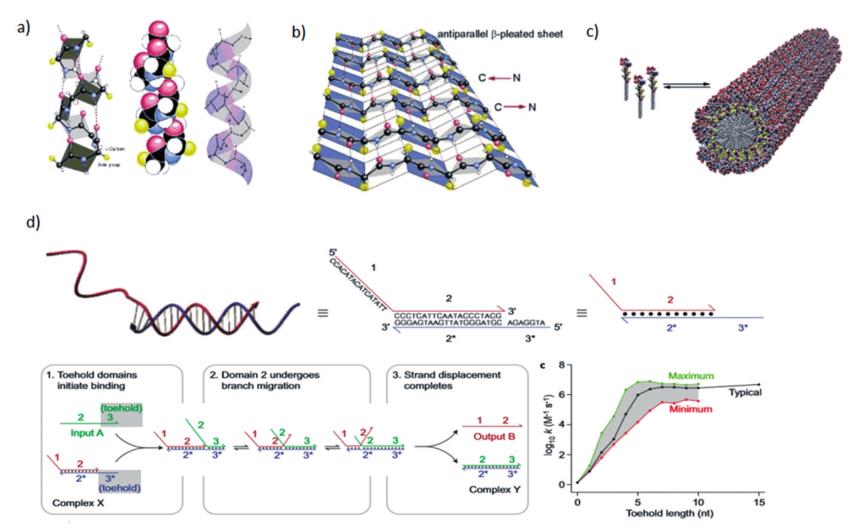
(a,b) Schematic diagram of the α-helix and β-sheet secondary structures of the peptides
(c) Schematic illustration showing the self-assembly of amphiphilic peptides
(d) Schematic diagram of the DNA chain displacement reaction and its dynamics
>> Omizzur Peptide : A company focus on custom peptide synthesis
Read Related Articles:
Copyright © 2020 Omizzur Inc | Terms & Conditions | Privacy Notice | Sitemap